Cotton Research
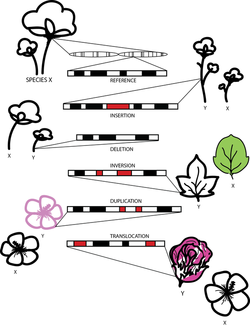 Schematic representation of genomic structural variations. Structural variations are large (>1kbp) rearrangements of DNA that frequently result in phenotypic differences. These variants include insertions, deletions, inversions, duplications, and translocations. By comparing genomes of different species, large chromosomal changes can be identified. Image by C. Evans
Schematic representation of genomic structural variations. Structural variations are large (>1kbp) rearrangements of DNA that frequently result in phenotypic differences. These variants include insertions, deletions, inversions, duplications, and translocations. By comparing genomes of different species, large chromosomal changes can be identified. Image by C. Evans
This project is done with Josh Udall at the Plant and Wildlife Science Department of Brigham Young University in Provo, UT
The cotton genus (Gossypium) is a diverse genus with approximately 50 species with eight monophyletic genome groups (A through G, plus K) from three continents. Approximately 1-2 MYA the African/Asian A-genome and an American D-genome species hybridized. From this single polyploidiztion event, eight polyploid species independently evolved in the Americas, two of which have been domesticated, and constitutes more than 95% of the worlds current cotton production.
To understand genome evolution of polyploid plants, physical maps of diploid and polyploid cotton genomes will be created. By doing so we will be able to uncover the structural variation between extant genomes, the structure of a polyploid common ancestor, and structural rearrangements that were fixed or occurred during allopatric speciation. Understanding the structural rearrangements of plant genomes will give us insights into speciation, phenotypic variability, and crop improvement.
To study structural variation, traditionally researchers have utilized techniques that first chop up genomic DNA into very small fragments losing structural relationships between regions. In this project, structural variation will be investigated using physical maps created using the Irys System from BioNano Genomics. The Irys System uses long DNA (high-molecular weight DNA) that remains intact. Analysis of large whole molecules to generate genome maps enables a direct view the context of elements in the genome and comprehensively catalog structural variation. See my review publication on Genome Mapping in Plant Comparative Genomics.
The cotton genus (Gossypium) is a diverse genus with approximately 50 species with eight monophyletic genome groups (A through G, plus K) from three continents. Approximately 1-2 MYA the African/Asian A-genome and an American D-genome species hybridized. From this single polyploidiztion event, eight polyploid species independently evolved in the Americas, two of which have been domesticated, and constitutes more than 95% of the worlds current cotton production.
To understand genome evolution of polyploid plants, physical maps of diploid and polyploid cotton genomes will be created. By doing so we will be able to uncover the structural variation between extant genomes, the structure of a polyploid common ancestor, and structural rearrangements that were fixed or occurred during allopatric speciation. Understanding the structural rearrangements of plant genomes will give us insights into speciation, phenotypic variability, and crop improvement.
To study structural variation, traditionally researchers have utilized techniques that first chop up genomic DNA into very small fragments losing structural relationships between regions. In this project, structural variation will be investigated using physical maps created using the Irys System from BioNano Genomics. The Irys System uses long DNA (high-molecular weight DNA) that remains intact. Analysis of large whole molecules to generate genome maps enables a direct view the context of elements in the genome and comprehensively catalog structural variation. See my review publication on Genome Mapping in Plant Comparative Genomics.
Orphan Crop Genomics
Through collaboration with scientist at BYU's PWS Orphaned Crop Lab, I contributed to the publication of the plastome of two orphaned crops. Orphan crops are underutilized species for providing food on a global scale and a typically not traded with internationally, but often play a major role in regional food security. In contrast maize, wheat, and rice account for a large portion of the world's consumption of calories. I worked on the Chloroplast genome in Amaranth and the Mitochondria and Chloroplast genome of Quinoa.
